The Dengue Virus Antibody Database is an online repository containing DENV-specific mAbs, annotated with information on 1) its origin, including the immunogen and host immune history, 2) binding or neutralization data against four DENV serotypes, and 3) epitope mapping at the domain or residue level.
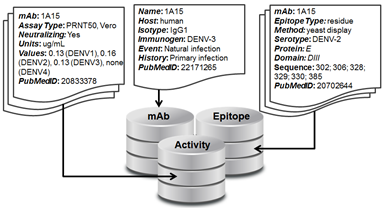
There are three linked sections of the database: mAb, Activity, and Epitope (see figure).
1. mAb - The section contains a single record for each mAb in the database that includes the mAb name and information about its origin, including the host organism, the isotype, the immunogen, the exposure event that lead to the antibody, the host immune history, the selection criteria used to isolate or select that mAb for study, and the PubMedID of the reference that first reported the mAb.
2. Activity - This section can contain multiple records for each mAb and includes information on the activity assay details and data against all four DENV serotypes, and the corresponding PubMedID for its reference. This data could be qualitative, such as in the case of a Western Blot, or quantitative, as in the case of titers from a neutralization assay.
Defining serotype specificity
We classified the serotype specificity of each mAb as ‘type,’ ‘sub-complex,’ and ‘complex’ based on the number of serotypes it was reactive towards. Complex Abs bind or neutralize all four serotypes, type-specific Abs bind or neutralize only a single serotype, and sub-complex Abs bind or neutralize more than one, but not all four serotypes.
3. Epitope – This section can contain multiple records for each mAb and includes information on the epitope resolution or type (‘residue’ or ‘domain’), the epitope mapping method, the domain of E that the mAb was mapped to, the E amino acid sequence that describes that epitope, and the PubMedID of the corresponding reference. Epitope resolution refers to the level of detail of a given method of epitope mapping – mutagenesis or x-ray crystallography gives residue-level information while western blot or yeast display gives domain-level information.
The database can be accessed publically at http://denvabdb.bhsai.org. The main page allows users to search mAbs in the database by mAb name, host, infection type, serotype specificity, and epitope mapping method. Selection of any of these fields results in displaying a subset of mAbs (‘selected mAbs’) in the database that fulfills the search criteria. A composite epitope map displays the combined epitopes of all selected mAbs is on the right. The corresponding Activity and Epitope records for the selected mAbs can be downloaded in .XLS, .PDF, .CSV, and .XML formats through links on the bottom of the search results.
Each antibody in the database has an Antibody page which provides all the associated Activity and Epitope records for that antibody along with visualization of the epitope residues mapped to the E protein structure and to the whole virus envelope structure.
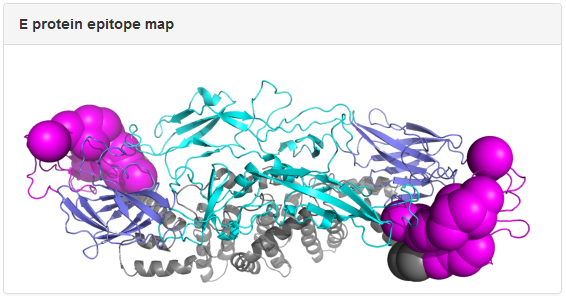
Example: mAb 3E31. The dengue E protein in the epitope map is colored by domain: slate, cyan, magenta, and gray for Domain I, Domain II, Domain III, and the transmembrane region, respectively. The spheres correspond to epitope residues identified in epitope mapping experiments, with the size of the spheres reflecting the resolution of the epitope mapping method. In the above example, epitope residues for 3E31 were mapped to Domain III of E protein from x-ray crystallography data.
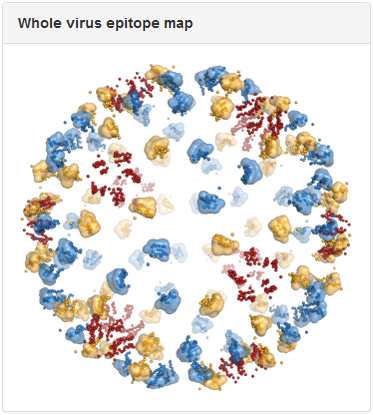
Above is the same antibody, 3E31, mapped to the whole virus envelope. Epitope residues identified through epitope mapping experiments are shown as spheres and colored red, yellow, and blue, to correspond to the three distinct symmetry-related epitope arrangements that result from the icosahedral geometry of the dengue virus envelope. We developed a computational method to predict quaternary epitopes on the virus envelope starting from experimentally determined epitope residues using criteria such as epitope compactness and structural similarity across symmetry environments. For antibodies with residue-level epitope information, computationally predicted quaternary epitopes are shown as surface ‘blobs’ surrounding the corresponding epitope residues. In the above example, quaternary epitopes were predicted in two of the three symmetry environments (blue and yellow). Although residues were mapped to the third symmetry environment (red), they failed to pass the criteria for quaternary epitope prediction.
Download all antibodies and corresponding activities and epitopes information.
Contact us at denvabdb@bhsai.org
Reference: Chaudhury, S., G. D. Gromowski, D. R. Ripoll, I. V. Khavrutskii, V. Desai, and A. Wallqvist. Dengue virus antibody database: systematically linking serotype-specificity with epitope mapping in dengue virus. PLOS Neglected Tropical Diseases. 2017 February 21; 11(2):e0005395. [PDF]